|
<< HOME
disentangler The disentangler (Kumasaka et al., 2011) is a visualization technique for linkage disequilibrium mapping and haplotype analysis of multiple multi-allelic genetic markers. The software was implemented by Java language with Java Runtime Environemnt (JRE) 1.5 or later. A prototype of this technique was first used in Okada et.al (2011) for the comperative association study of ulcerative colitis and Crohn's disease. Latest Version (2011-Oct-01): Executable-JAR | Example Data 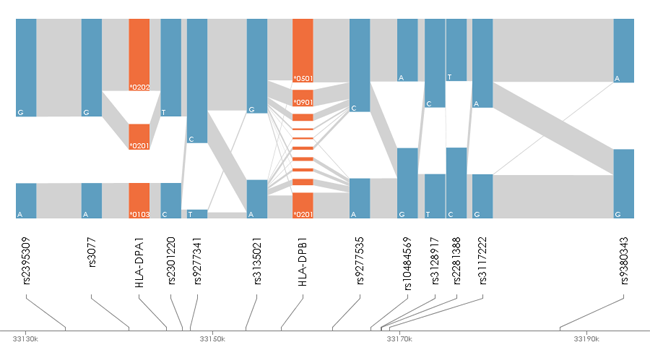 Before using the software, please make sure that the JRE 1.5 or later is installed in your computer. Then, download the executable-JAR file (disentangler.jar) above, and double-click on the file. You may also run the program from the computer prompt: % java -jar ./disentangler.jar Then you will see the program window onto which you may drag and drop your own data set. To run the disentangler with a large scale data set, enter the following command with <Mb> mega-byte memory at the computer prompt: % java -Xmx<Mb>m -jar ./disentangler.jar PREPARE INPUT FILE 1. General Our software requires only a flat file (.txt file) with header in general. The file should be 'TAB' separated, in which each column corresponds to each marker. The column is further separated by 'SPACE' to specify two alleles derived from parents. The allele can be any character string (e.g., "*1234", "A", "007",...), but without 'SPACE' and capital 'N' (assigned to the missing value). In this regard, any SNP genotype can be incorporated as a pair of two alleles (e.g., "A B", "C C", "T G", etc.) without loss of generality. Again, the capital 'N' cannot be used for a specific allele but for the missing allele. The following figure shows an example of the input file format: 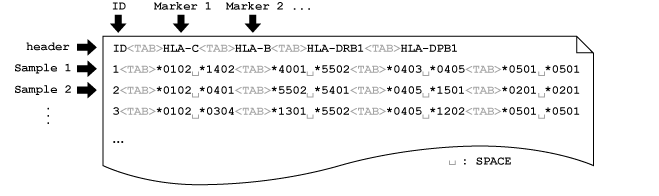 The leftmost column (the first column) may be the sample ID, each of which must specify the unique row (any repeat of the same ID is prohibited). Note that the ID column can also be omitted. The missing genotype can by represented by "N N","- -", "", "NA" and "-". If a part of the genotype is missing, you may specify a combination of the allele and a missing value specified by either "N", "-" or "" (e.g., "*3029 -", "A N", "B ", etc.). 2. Information file (optional) If you want to introduce chromosomal positions of each markers on the display, you may introduce an additional information file (.info) for convenience. The information file is also 'TAB' separated, and the first column indicates the marker ID and the second column indicates the chromosomal position (see the following figure). 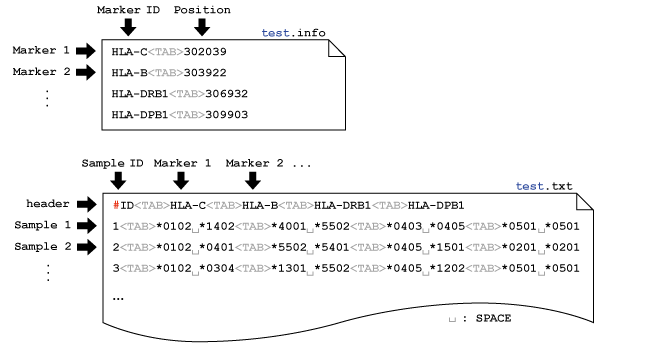 Here the header of the data file must be removed or comment out by using "#". Note that the "#" (comment-out) can also be used at any row to get rid of the subject from the data. In addition, the file name except for the suffix (.info) must be the same as that of the main data file (test.info and test.txt in this example). 3. SNPs and CNPs Not only multi-allelic markers but also single-nucleotide polymorphisms (SNPs), copy-number polymorphisms (CNPs), allele-specific CNPs or mixture of these can be displayed on a single display. Here the marker type (variant type) should be specified on the third column of the information file (*.info). The following example shows that a non-allelic CNP and an allele-specific CNP are followed by the same SNP marker with different representations. 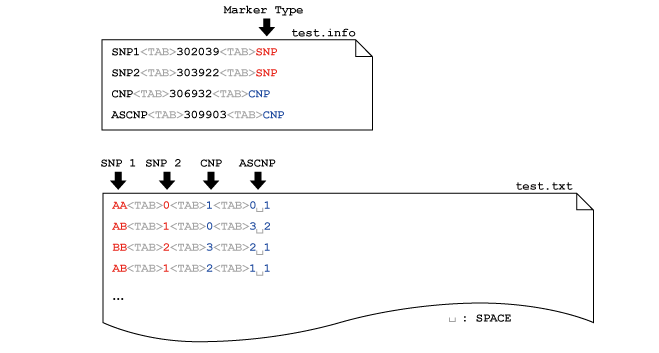 Each SNP genotype can be encoded as a pair of two alleles without 'SPACE' (e.g., [AA/AB/BB], [CC/CT/TT], etc.) or the major (or minor) allele count of [0/1/2]. Similary, non-allelic CNP can be specified by non-negative integer of genotypic copy number [0-Inf]. The allele-specific (biallelic) CNP can also be specified by a pair of two numbers with 'SPACE' in between. Each of the two numbers indicates the number of copies of each of the two alleles. Limitation: the current disentangler can accomodate only biallelic CNPs, that is, the genotype consists of multiple copies of just one or two alleles (e.g., "AAA", "ABB", "AAAAABB", etc.). GUI Several interactive user interfaces are implemented so far. The user can click either an allele or a haplotype between two alleles on the display to see conditional distributions of haplotypes that involve the specified allele(s). REFERENCES N. Kumasaka, Y. Okada, A. Takahashi, M. Kubo, Y. Nakamura and N. Kamatani. Disentangler: a visualization technique for linkage disequilibrium mapping using multi-allelic loci ; (Abstract / Program #708F). Presented at the 12th International Congress of Human Genetics/61st Annual Meeting of The American Society of Human Genetics, October 14, 2011, Montreal, Canada. Okada Y et.al. (2011) HLA-Cw*1202-B*5201-DRB1*1502 haplotype Increases risk for ulcerative colitis but reduces risk for Crohnfs disease, Gastroenterology 141(3) 864-71. << HOME |